
Can architects help create next-generation treatments for cancer and lung disease? Will the buildings of tomorrow have intelligent skins? What does figure skating have to do with it? An unusual partnership between Penn cell biologists and design students is tackling a lot of strange questions. Their answers may rewrite the rules of biomedical research.
By Trey Popp | Photograph by Candace diCarlo
Eight months after coming to Penn to pursue a master’s degree in architecture, Erica Swesey Savig began leading a second life. While her fellow students papered their workstations with maps of city intersections, she would slip out of Meyerson Hall and walk across 34th Street to the Vagelos Laboratories. Inside she would don a pair of pale green latex gloves. If you happened to drop by on the right day, you’d then find her filling a sterile tissue-culture dish with collagen gel and a cloned line of vascular smooth muscle cells derived from a rat embryo.
Magnified in the climate-controlled chamber of a Nikon biological microscope, the cells would seethe and swarm in an apparent chaos of movement. Savig would watch. Every five minutes she would snap a digital photograph. Finally she would import several hours’ worth of images into a three-dimensional modeling software called Rhino that is most commonly used in industrial design.
Then the interesting part would begin. Depending on the characteristics of the collagen gel she used, these genetically identical cells appeared to move in different ways. Savig wanted to answer a deceptively simple question: Was it possible to distinguish between different environmental conditions by looking at cell movement alone?
Savig is not a scientist. Before that first foray into the lab in April 2007, she wanted to design buildings. The story of how cell biology commandeered her attention arises from one of the most unusual interdisciplinary collaborations to emerge at Penn in recent memory. It has enlisted architecture students in a quest to open new fronts in the treatment of lung disease and breast cancer, and aims to equip designers with tools that can bring their field into what some futurists have dubbed the Age of Biology.
To be more specific, Erica Savig has spent the last year and a half looking at smooth vascular muscle cells for two reasons. The most immediate is that they constitute a model of what happens in pulmonary arterial hypertension, a fatal disease whose causes are poorly understood. If she can leverage the unique capabilities of her architectural tool set to shed light on the derangement of tissue in diseased lungs, it could pave the way for a completely new approach to diagnosis and treatment.
Yet it is no coincidence that she is examining a cellular system that bends its behavior in accordance with the environment that surrounds and permeates it. Savig wants to incorporate the same sort of dynamic process into a building façade, enabling it to “intelligently” adapt to environmental conditions as they change—for instance, by varying the amount of sunlight or heat that can pass from the exterior to the inside. Marrying her laboratory insights with digital algorithms and nanoscale materials science, she aims to build a small physical prototype this year.
“I don’t want to design buildings anymore,” she said over the summer with a laugh that was only halfway in jest, as she reflected on the unexpected trajectory of her architectural education. “I want to design something that behaves.”

It was a dry November day in 2005 and Peter Lloyd Jones had wandered out from his usual turf in the Vagelos Laboratories, where he makes his home within the Institute for Medicine and Engineering (IME). Jones is an associate professor of pathology and laboratory medicine, and currently directs the Center for Pulmonary Arterial Hypertension Research. If his professional titles suggest a double dose of introversion, he resolutely plays against stereotype. In his office there is an Oscar Wilde action figure propped up against a wall-mounted marker board. The Irish playwright’s penchant for wit often finds a reflection in the British pathologist’s banter. “My favorite quote,” Jones told me, “is: ‘A true friend stabs you in the front.’”
Jones can’t remember exactly where he was going that day in November, but his unexpected detour remains vivid. “I think I was walking to get some coffee, and there was a sign: Non-Linear Systems Organization,” he recalls. “And I thought, I’m a non-linear systems biologist. I’m going to walk in.”
The NSO is a research group that had recently been started within the School of Design under the leadership of Cecil Balmond, an internationally renowned structural engineer and the Paul Philippe Cret Practice Professor of Architecture. Its first annual conference was taking place in Meyerson Hall. Most of the participants hailed from university architecture departments around the country, plus the odd mathematician, engineer, and software designer. They had gathered to address questions like: “How can scientific models of complex phenomena in mathematics, nature, and the universe be most effectively employed in the design and fabrication of structures for human life and enjoyment?”
Jones wandered in and was “completely blown away” by what he heard. There weren’t any biologists among the speakers and panel moderators, but some of the ideas they were batting around evoked striking parallels to his own work.
Like an increasing number of his colleagues in the life sciences, Jones has entered what is sometimes called the postgenomic era of biological research. Before the sequencing of the human genome was completed in 2000, the reigning assumption among molecular biologists was that each protein manufactured by our bodies derived from a unique corresponding gene, and that the destiny of a given cell was driven by the genetic code it carried. But the data that came out of the Human Genome Project told a different and much more complicated story. Our bodies make some 90,000 distinct proteins—the chief actors within cells—from a mere 30,000 genes. Furthermore, evidence is accumulating that a cell’s local environment can exert a dominant influence over gene expression—which can in turn impact that very same microenvironment. Understanding these non-linear, dynamic feedback loops has become one of the major challenges of contemporary biomedical research.
What this means in terms of human health and disease is explained by Anne Plant, a biochemist at the National Institute of Standards and Technology in Washington. “It’s becoming more and more appreciated,” she says, “that if you put cells in one kind of extracellular-matrix environment, or another extracellular-matrix environment, and treat them otherwise exactly the same—with the same chemical stimulants or the same pharmaceutical agents—you will get different responses.”
The average high-school biology teacher probably doesn’t spend much time on the extracellular matrix, but animal life would be all but impossible without it. The ECM is the connective tissue that provides structural support to living cells, giving them a sort of scaffolding to which they can anchor. It also regulates communication between cells, stores and releases chemicals that can trigger a range of cellular functions, and governs the movement and migration of cells through its intricate architecture. The complexity of the system beggars description. The components of extracellular matrices are manufactured inside of their resident cells, which then fall subject to the influence of structural and biochemical changes within the scaffolds they have excreted.

Models by Sabin, Wang, and Jones; photo by Candace diCarlo
The far-reaching influence of the ECM has profound implications. “You could potentially treat diseases and cause cell behavior to change by changing the extracellular matrix’s elasticity,” for example, says outgoing IME director Peter Davies, the Robinette Foundation Professor of Cardiovascular Medicine and a professor of bioengineering as well as pathology and laboratory medicine. “And in fact, in breast cancer it works,” he says. “If you change the matrix’s physical properties, you can cause an epithelial tumor in culture to reorganize back to its normal form—rather than being a cancerous cell which grows all over the place. And its metastatic potential declines precipitously.
“You could imagine,” Davies continues, “that instead of—or in addition to—treating a breast tumor with chemotherapy or radiotherapy, you could also alter the environment around the cell by micro-injecting a gel of some kind, or a matrix with properties that you know favor the maintenance of normal.”
In other words, differences or changes in tissue architecture can have major repercussions on the development and treatment of disease. This “radical but true notion” was already apparent to Jones when he walked into the NSO conference. But he walked out with a novel idea: maybe architects would be able to look at the tissue systems in his lab and see things that he was overlooking, or didn’t know how to analyze. The person who would help him shape and realize this vision was NSO founding member Jenny Sabin, a lecturer in the School of Design.

Jenny Sabin is the kind of teacher whose main effect on students is probably either to bewilder them or change them forever. She speaks in trenchant paragraphs that occasionally threaten to buckle under the pressure of the ideas crammed inside of them. “We’ve talked about the cell as a loom,” she said one day over lunch, drawing an analogy between cell behavior and textile fabrication. “There’s this incredible thing going on where the cell is not only weaving its own environment, it’s moving within the environment and responding to it. And a lot of my former work was looking at the loop that exists between computation, material, weaving, and other textile processes. And I see parallel—different, but similar—loops in the research Peter is doing.” Her expertise in textile structures and computation imbues her design sensibility with a rigor that can be alternatively daunting or inspiring.
Sabin and Jones spent the next year discussing how they could collaborate. They had their sights set on something more concrete than a simple exercise in cross-disciplinary creative fertilization. Jones wanted to bring a different mentality into his lab, one that hadn’t been shaped by the sort of “enclave thinking” that can blinker scientists ensconced in a narrow specialty. Sabin quickly saw that this would involve problem-solving challenges that were salient to her field. “We would still be working with design,” she says, “but at a radically different scale.”
At Penn, the architecture curriculum extends a fair piece beyond the activities that typify the average practicing firm. “In a very short period of time, we give people an awful lot of skills,” says Professor Detlef Mertins. “We develop a sort of hand-eye coordination, we develop their imagination, we develop lateral thinking—and we do that through a whole variety of exercises.”
Some of those exercises strike out into pretty abstract terrain, but Mertins, who has written extensively about the intersection of biology and architecture, suggests that using Jones’ lab as a kind of classroom isn’t that far-out. “We’re interested in exploring how certain things that they’re studying in terms of cells—the surface of cells, the communication between the inside and outside of cells—can inform our way of designing building envelopes, or roofs, or canopies, or components of buildings.
“Buildings are shelters that separate the inside from the outside,” he continues, “but they also allow all kinds of communication between the inside and the outside through the envelope—the skin of the building, if you like. Whether it’s heat gain through windows, or ventilation, or other kinds of things, there’s an interchange that goes on between the inside and the outside. And of course these days, we want to optimize energy consumption … so at a time when all these environmental issues are so strongly in the foreground, it’s a very healthy thing to be exploring the potentials of natural models for all the aspects we deal with in buildings.”
In 2007, with substantial if cautious support from their respective departments, Jones and Sabin formed LabStudio. Three postdoctoral and graduate students in the IME were each paired with a counterpart from the School of Design. Jones and Sabin gave each team a different research brief. One would focus on cell surface design, another would investigate cell networking behavior, and the third—where Erica Savig landed—would concentrate on cell motility.
“The advantage of formulating these scientist-architect pairings,” Sabin says, “was that experiments could be redesigned based on the architect’s objective observations, intuition, and requests—and new tools could be developed and modified based on the scientist’s specific hypotheses.”
Jones and Sabin also created and co-taught a class called “LabStudio: NonLinear Design Diagnostics and Tooling.” The last word of that title is in many respects at the heart of their joint endeavor.
When Erica Savig peered through a microscope lens at smooth muscle cells, she wasn’t doing anything that numberless scientists haven’t done before. Cell motility has fascinated biologists ever since Antonie van Leeuwenhoek discovered the “pleasing and nimble” motions of single-cell organisms scooting about in rainwater in 1675. But when Savig imported those digital images into software that had been developed for architects and industrial designers, that was something new.
“Architects, it turns out, have incredibly sophisticated tools for visualizing things in four dimensions—three-dimensional space across time—and have incredibly sophisticated mathematical tools for dealing with spatial things,” says Mark Tykocinski, who was the Simon Flexner Professor and chair of the Department of Pathology and Laboratory Medicine before becoming dean of Jefferson Medical College in December. “Those things haven’t informed medicine and biomedical research in a very serious way until now.”
The notion that state-of-the-art biomedical labs have been stuck with tools that would get laughed out of an architecture studio is bound to surprise anyone who compares the typical operating budget in each realm. But there is truth in it, and consequences. In a paper slated for the January 2009 issue of the Journal of Mathematical Biology, University of California cell biologist and mathematician Alex Mogilner writes that “lack of standard modeling methods, difficulties in translating biological phenomena into mathematical language, and differences in biological and mathematical mentalities continue to hinder the scientific progress.” The title of his article is phrased as a question: “Mathematics of Cell Motility: Have We Got Its Number?” Three centuries after van Leeuwenhoek set biologists on the hunt, Mogilner’s answer, effectively, is no.
Peter Jones’ question was whether architects could help to push the ball forward at a faster clip. A year and a half later, he was having trouble containing his enthusiasm. “I mean, we’ve seen things that nobody’s seen before! You know, we’ve been sitting on some of this data—looking at these images, looking at these moving cells, looking at these tissues—and didn’t see what they can see.”
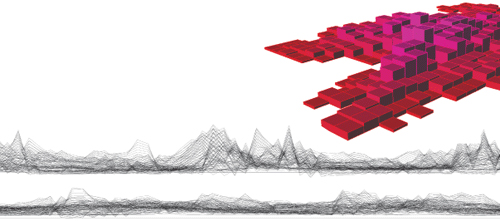
In science, it is one thing to see, and another to be able to quantify. By dint of her spatial acuity and her use of unorthodox software tools, Erica Savig has accomplished the former. But the latter is where the real test lies. She is currently tackling it with Mathieu Tamby, a postdoctoral cell biologist who came to the IME last year.
In August, Savig and Tamby presented some of their research at an informal lab review. They showed still photographs and reconstructed videos of smooth vascular muscle cells under two conditions: some had been seeded in native collagen, which approximates a healthy extracellular matrix; others had been seeded in denatured collagen, which models the ECM’s deterioration in pulmonary arterial hypertension. Collagen is the most abundant protein in mammals and a major component of the extracellular matrix.
The pictures told a more or less straightforward story. As Savig had put it earlier, “You see already the cells are behaving differently. But how do you quantify this?”
Tamby cued a slide depicting some of the standard ways cell biologists characterize the behaviors he had just projected onto the screen. “These are classical ways of representing these parameters with numbers,” he said. “So we look at cell velocity on native and denatured collagen, and this was looking at the average velocity of one cell.”
Tamby’s graphs looked basically the same to an untrained eye—or to a trained one, for that matter.
“So this classical way of viewing numerical data isn’t quite capturing differences in behaviors that we’re seeing,” Savig interjected.
“But in science, we like numbers,” Tamby responded. “We like to obtain significant differences.”
For something as simple as average cell velocity, the methodology for making statistically significant comparisons is relatively straightforward. But what seemed to differentiate the two cases in question was occurring at a finer level of detail. Cells on native collagen sometimes appeared to form different patterns of alignment than cells on denatured collagen. Or they bent themselves into what looked like different sorts of shapes, which Savig would describe during brainstorming sessions as “scouts” and “oafs” and “middlemen”—sometimes sending Tamby, whose mother tongue is French, scrambling for the Web-based translator bookmarked on his browser.
Savig turned to the Rhino 3-D modeling software to translate some of these concepts into geometrical data. “Really, what we try to do as architects is take a lot of information and almost reduce it to a series of spatial relationships,” she says. “And in essence, that’s what biologists are trying to do as well. They’re trying to look at stuff and come up with some kind of relationship or rule set that explains what’s happening. And these digital techniques we use in architecture help us to reduce the information, filter through it, and find these relationships.”
The goal was to translate subtle differences in those spatial relationships into unique signatures betraying whether the underlying collagen mimicked a healthy or diseased extracellular matrix. If that was possible, it could spur a huge advance in pulmonary arterial hypertension diagnosis and treatment.
Pulmonary arterial hypertension (PAH) involves the progressive derangement of blood vessels in the lung, which overwhelms and exhausts the heart until it fails. There is evidence suggesting that PAH actually encompasses multiple diseases. At present, however, it is so difficult to distinguish between them that doctors effectively choose a drug treatment through guesswork. The diagnostic tools they use are primitive. “Right now, we have these catheters that give you pressure tracings, and X-rays that can show you two-dimensional and sometimes reconstructed three-dimensional images of what their lungs look like,” says Darren Taichman, the associate director of Penn’s Pulmonary Vascular Disease Program. “But what that doesn’t do for me is tell me very much about who should be treated which way.”
The FDA has approved six new drugs for PAH in a relatively short span, but there is a huge variation in how patients respond to them. “These drugs are toxic, like any other drug, and they’re unbelievably expensive,” Taichman says. “And when you’ve got a disease that kills people relatively quickly—and more recent data which suggest that the sooner we get people on therapy, the better they’ll do in the long run—it would really be nice if you could choose the right therapy sooner.”
What Taichman and Jones are searching for is a way to zero in on what’s happening on the cellular level. They think that whatever drives the cell changes which lead to blood-vessel derangement might be reflected in the general bloodstream. If so, it might be possible to add Savig’s smooth vascular muscle cells to a patient’s blood-tissue sample rather than collagen, and refine the diagnostic method to derive personalized signatures for PAH patients—which in turn could help doctors determine which treatment would likely be most effective.
Traditional clinical diagnostics revolves around detecting particular molecular substances in various body fluids or tissues. But the problem in a complex nonlinear system is that “there is no single molecule that really controls everything,” Peter Davies points out. “In fact, there are many biomarkers, many principal players, and the principal player at one time may be—a day later or an hour later or a minute later—no longer the principal player.” Jones has spent much of his career investigating the role of particular molecules in PAH—for instance, an ECM protein called tenascin-C that is critical in the formation of blood vessels in the lung. The LabStudio collaboration opened the possibility for a different strategy.
“There is a whole other realm of potential diagnostic tests that revolve around what you might call functional assays, or functional readouts,” says pathologist Mark Tykocinski. “So instead of trying to catalogue all those molecular drivers, you look at the other end: you look at the cell that’s affected by all those molecular drivers. In fact, you don’t even have to know all the molecular drivers—you just look at the functional readout.” Some functional readouts are easy to read, but the intricate choreography of cell movement is likely to overwhelm even a microscope-aided eye. “So having new tools to quantitatively analyze something like cell motility, that’s where the link to architecture weighs in.”
Looking at the smooth muscle cells, it had seemed to Savig (and Jones) that the movements of filopodia—slender projections of cytoplasma that extend from the leading edge of a cell wall—were important. So she resolved to trace the edges of cells in Rhino, creating a digital data set that she could manipulate and hopefully deduce rules from. But a big obstacle stood in the way: there was no system of mapping or mathematical coordinates that easily lent itself to measuring those wild, horn-shaped protrusions.
When Jones and Jenny Sabin initiated their partnership, they hoped that injecting an outsider’s mentality into the lab “would lead to both extraction and abstraction of new biological information.” That was exactly what Erica Savig was now on the cusp of doing. It would be hard to overstate the peculiarity of the tack she chose. She turned to a resource that no scientist in the English—or French—speaking world would ever have thought to consider, even after exhausting a thousand other ideas. It was a paper written in 2006 by a student of Sabin’s named Jackie Wong GAr’07. Presumably, even Wong himself never imagined that his work might have any relevance to cell biology. Titled “Dance and Space,” it was an analysis of the spatial properties of figure skating—or, to be more specific, the compulsory ice dancing event.
“The project tracked the movement of ice dancers,” Savig explains. “But not just how they moved together along a plane: also the movement of their hands and legs”—which are, in a sense, a type of bodily protrusion not that different from a cell’s filopodia.
She adapted some of Wong’s ideas to develop her own system for characterizing the irregular motion of cell shapes. More importantly, she and Tamby have been able to do so with sufficient mathematical rigor to convert some—though still not all—of the behavioral differences between their two experimental conditions into hard numerical data. Yet that leads to a further challenge. A tiny bit of tissue can yield vast quantities of data. So much, in fact, that it becomes hard to figure out how to handle it. But that too was why the architects had come into the lab.

“We are approaching an era of these way-beyond-gigabyte quantities of data from individual patients or cells,” Peter Jones says. “How the heck can you possibly use traditional methods to number-crunch and display those data? You probably can’t. So can we do it by looking at form and patterns?”
What design students bring to LabStudio’s table are the skills to winnow and sift through the vast mountains of information to bring relevant patterns to the surface. That may sound hopelessly abstract, but in a sense, it is an architect’s most basic job description. “Design is an act of filtering,” Sabin says. A building begins as a list of demands and constraints that essentially takes the form of raw data: the lot size, the number of square feet needed for a variety of activities, the budget, the cost of materials, zoning restrictions on height and shape, fluctuating plumbing and lighting and energy needs … and on and on. “You’re constantly navigating all of these different constraints and parameters that make up a project, because in the end, the design is a synthesis of all of them.”
As the list of parameters has grown to encompass an increasingly complex set of requirements—everything from minimizing construction waste to maximizing a building exterior’s capacity to channel air flow toward roof-mounted wind turbines—digital design tools have changed radically.
“You may think that designing with a computer involves using a mouse,” Detlef Mertins says. “And it does, if you’re using AutoCAD and other conventional software. But it’s also possible to design by writing script or code—that is to say, you do it numerically through functions and algorithms.”
The most recognizable product of this approach is probably Beijing’s National Aquatics Center, where Michael Phelps hauled in his eight gold medals at the 2008 Olympic Games. The Watercube derives its iconic façade from a set of mathematical equations describing the structure of soap bubbles. Each of the building’s four walls is nearly as long as two football fields, yet they alone support the gigantic, 7-acre roof. “Over such a wide span of column-free space, the need to minimize the self-weight of the structure is paramount, as most of the structural work involves ensuring the roof can hold itself up,” architect and writer Michael Weinstock observed in the journal Architecture Design. The laws of gravity and torsion, in other words, become tricky parameters indeed. So the shapes and locations and thickness of the 4,000 weight-bearing “bubbles” that make up the Watercube’s structure were determined by the meticulous refinement of digital scripts and algorithms—which can be converted seamlessly into factory instructions. “At the scale of very large architectural projects,” Weinstock continues, this process “becomes not only the significant design strategy, but also the only economic means of reducing design data for manufacturing.”
In an analogous way, Savig applies scripts—albeit “simple” ones, she stresses—to her data in Rhino. Only rather than deriving a structure, she is creating novel graphic representations that make relevant patterns legible.
What Peter Jones wanted, and Jenny Sabin hoped to provide along with her students, was a new way of seeing. They are starting to get it.
“This isn’t immediately fundable,” Jones says. “But it’s starting to move toward some sort of acceptance. If we’d put this in two years ago to the NIH, it would have been a joke to them. Now they’re crying out for new modes of visualization. And so is the NSF. So there’s a paradigm-turn occurring, I think.”
In some ways, it is a paradigm shift that the art world has anticipated. Over the summer Sabin exhibited a different LabStudio product at SIGGRAPH, an annual exhibition of computer graphics and interactive design that features the work of world-renowned architects like Frank Gehry and Zaha Hadid. The piece, “Branching Morphogenesis,” is an abstract sculptural rendering of lung endothelial cells interacting within a three-dimensional matrix environment. Made from some 75,000 color-coded cable zip ties, it comprises five interwoven curtains 12 feet high, 15 feet wide, and eight feet in depth. The curator of the Ars Electronica Museum in Linz, Austria, recently selected it for an exhibition celebrating that city’s stint as the 2009 European Capital of Culture.
“It was interesting when Peter Davies walked in,” Jones recalls of his colleague at the Institute for Medicine and Engineering. “I think we’d put up three sheets that day, and he immediately identified each component as a data point.”
“The thing that struck me,” Davies says, “is you had rows and layers, and each of those tags of the tapestry was an interpreted unit of information coming from the cells … And while we were there, one of the art students projected light through it, so that it threw an image onto the wall behind the curtain—and that’s another derivative, because you are turning a three-dimensional model into a two-dimensional image!” The passage of time has not blunted Davies’ intellectual high. “And I was getting ahead of myself, saying, so now you can gain further information by manipulating that three-dimensional model … It starts stimulating you to think about different stratagems for taking the derivative of huge amounts of biological information.”
Or as Jones puts it with a chuckle, “The data become something very different when you’re walking amidst it.”

There is a paradox buried in the contemporary practice of science, and biomedical researchers may well be the poster children for it. They thrive on innovation but abhor risk. Grant funding is the engine of every academic lab, and wild ideas usually don’t pay. There were no professional incentives for Peter Jones to hire architecture grad students into his lab, and plenty of reasons that would have persuaded a by-the-numbers biologist to abandon the notion. By the same token, an architect seduced by living tissue makes a gamble of her own.
“We’ve both faced some ridicule, I must say,” Jones sighs, drawing a nod of agreement from Sabin. “There’s a perception of artists as flakes and scientists as geeks, and never the twain should meet. And there are certain places I’ve presented where it’s considered to be sort of play, rather than product.”
Yet it’s hard not to notice the way their collaboration has crept into the minds of an awful lot of their colleagues—at Penn and elsewhere. “We’re not really sure what’s going to come out of it,” says Anne Plant, who is confronting many of the same challenges as the leader of NIST’s Cell & Tissue Measurements Group. “One of the very exciting things about this work is that it’s very early, and very exploratory, and that’s what’s important … They are asking a question in a very different way than how it’s been asked before.”
Even if the exercise were to end today, LabStudio appears to have permanently enriched Jones’ basic research on breast cancer and lung disease. “There isn’t a single aspect of it now that hasn’t been touched by this,” he says. “It’s interwoven at this point.”
The feasibility of an adaptive building skin is yet to be determined, but for Sabin and many of her colleagues at PennDesign, that is only one among many potential benefits of a closer engagement with systems biology. “The modeling tools we’ve been using in design have become increasingly sophisticated—to the point where frequently you see the tools usurping the designers’ ability to navigate them effectively,” Sabin observes. The rigor that comes with plumbing cellular systems for lessons in how to apply next-generation tools with greater power and elegance should serve architects well.
“Crossovers are almost a kind of occupational hazard in architecture,” says Detlef Mertins. “And that’s a tradition that goes right back to the Renaissance. Architects were people who were effectively involved in military fortifications, engineering, fireworks—as well as building cathedrals and houses and other things.
“Architecture students who learn more about biology should be better able to understand and design buildings and environments within an ecosystems framework,” he continues. “To see buildings not as fixed, but as dynamic constructions—and as the result of processes that operate in time and involve energy and the transformation of material. The time frame may be decades or even centuries, but increasingly buildings—and certainly interiors—change within a few years.”
“And what is pathology?” Jones asks when he hears architects talking that way. “For me, it is the generation and loss of form.”
Now he’s sitting with Sabin, and the two are feeding off each other. The conversation ricochets from the basement membrane in breast cancer to deployable structures modeled after cell cytoskeletons, from high-throughput diagnostics to rapid manufacturing. Not for the first time, it is hard to keep up. LabStudio has spawned its own kind of data overload, a multitude of projects that will have to be winnowed to distill just one story. But then Sabin pauses.
“It was serendipitous that we met—” she says.
“—But no coincidence that we collaborate,” says Jones.
“Absolutely.”
“We’re still sending each other e-mails at four in the morning.”
But now the conversation is regaining speed, and four in the morning is too long to wait.

